cd /save_projet/eggtomeat/
mkdir RAW_DATA
mkdir RAW_DATA/RUN1
mkdir RAW_DATA/RUN2
mkdir RAW_DATA/RUN3
# Rename files
cd EGTM\ run\ 1_dec22/
rm -f Undetermined_S0_L001_R*
for i in *_R1_*.fastq.gz ; do id=$(echo $i |cut -d '_' -f 1) ; cp $i ../RAW_DATA/RUN1/${id}_R1.fastq.gz ; done
for i in *_R2_*.fastq.gz ; do id=$(echo $i |cut -d '_' -f 1) ; cp $i ../RAW_DATA/RUN1/${id}_R2.fastq.gz ; done
cd ../EGTM\ run\ 2_dec22/
rm -f Undetermined_S0_L001_R*
for i in *_R1_*.fastq.gz ; do id=$(echo $i |cut -d '_' -f 1) ; cp $i ../RAW_DATA/RUN2/${id}_R1.fastq.gz ; done
for i in *_R2_*.fastq.gz ; do id=$(echo $i |cut -d '_' -f 1) ; cp $i ../RAW_DATA/RUN2/${id}_R2.fastq.gz ; done
cd ../EGTM\ run\ 3_jan23/
for i in *_R1_*.fastq.gz ; do id=$(echo $i |cut -d '_' -f 1) ; cp $i ../RAW_DATA/RUN3/${id}_R1.fastq.gz ; done
for i in *_R2_*.fastq.gz ; do id=$(echo $i |cut -d '_' -f 1) ; cp $i ../RAW_DATA/RUN3/${id}_R2.fastq.gz ; done
cd /save_projet/eggtomeat/RAW_DATA/RUN1
mv H2O_R1.fastq.gz H2ORUN1_R1.fastq.gz
mv H2O_R2.fastq.gz H2ORUN1_R2.fastq.gz
mv H2Ob_R1.fastq.gz H2ObRUN1_R1.fastq.gz
mv H2Ob_R2.fastq.gz H2ObRUN1_R2.fastq.gz
cd /save_projet/eggtomeat/RAW_DATA/RUN2
mv H2O_R1.fastq.gz H2ORUN2_R1.fastq.gz
mv H2O_R2.fastq.gz H2ORUN2_R2.fastq.gz
mv H2Obis_R1.fastq.gz H2ObisRUN2_R1.fastq.gz
mv H2Obis_R2.fastq.gz H2ObisRUN2_R2.fastq.gz
cd /save_projet/eggtomeat/RAW_DATA/RUN3
mv H2O_R1.fastq.gz H2ORUN3_R1.fastq.gz
mv H2O_R2.fastq.gz H2ORUN3_R2.fastq.gz
mv H2Oter_R1.fastq.gz H2OterRUN3_R1.fastq.gz
mv H2Oter_R2.fastq.gz H2OterRUN3_R2.fastq.gz
mv H2Obis_R1.fastq.gz H2ObisRUN3_R1.fastq.gz
mv H2Obis_R2.fastq.gz H2ObisRUN3_R2.fastq.gz
mv H2O4_R1.fastq.gz H2O4RUN3_R1.fastq.gz
mv H2O4_R2.fastq.gz H2O4RUN3_R2.fastq.gz
cd ../RUN1
tar zcvf EGTM_run2.tar.gz *.fastq.gz
cd ../RUN2
tar zcvf EGTM_run2.tar.gz *.fastq.gz
cd ../RUN3
tar zcvf EGTM_run3.tar.gz *.fastq.gzEGGTOMEAT
Evaluation of the impact of chicken farming practices on microbial flux ( feces, cecal contents and carcasses)
This document is a report of the analyses performed. You will find all the code used to analyze these data. The version of the tools (maybe in code chunks) and their references are indicated, for questions of reproducibility.
Aim of the project
The aims of these analyses are to build amplicon sequence variants (ASVs) from raw reads and to affiliate ASVs sequences to obtain the taxonomic composition of the samples. Reads were provided by the @BRIDGE from an Illumina Miseq instrument (2x251 bp). The targeted amplicon is the V3-V4 region of the 16S rRNA. 3 different runs were performed.
Data management
All data is managed by the migale facility for the duration of the project. Once the project is over, the Migale facility does not keep your data. We will provide you with the raw data and associated metadata that will be deposited on public repositories before the results are used. We can guide you in the submission process. We will then decide which files to keep, knowing that this report will also be provided to you and that the analyses can be replayed if needed.
Sequencing data
Data were downloaded from Filesender, then deposited on the Front server (Bruyères-le-Châtel) and a copy was sent to the abaca server (Toulouse datacenter). First, we renamed FASTQ files to remove useless informations in filenames and correct duplicated ids.
# seqkit
cd /work_projet/eggtomeat
mkdir RUN1 RUN2 RUN3
qsub -cwd -V -N seqkit -q maiage.q -pe thread 4 -R y -b y "conda activate seqkit-2.0.0 && seqkit stats /save_projet/eggtomeat/RAW_DATA/RUN1/*.fastq.gz -j 4 > RUN1/raw_data.infos && conda deactivate"
qsub -cwd -V -N seqkit -q maiage.q -pe thread 4 -R y -b y "conda activate seqkit-2.0.0 && seqkit stats /save_projet/eggtomeat/RAW_DATA/RUN2/*.fastq.gz -j 4 > RUN2/raw_data.infos && conda deactivate"
qsub -cwd -V -N seqkit -q maiage.q -pe thread 4 -R y -b y "conda activate seqkit-2.0.0 && seqkit stats /save_projet/eggtomeat/RAW_DATA/RUN3/*.fastq.gz -j 4 > RUN3/raw_data.infos && conda deactivate"We can plot and display the number of reads (Figure 1) to see if the amount of reads by sample and if sequencing depth is homegeneous.
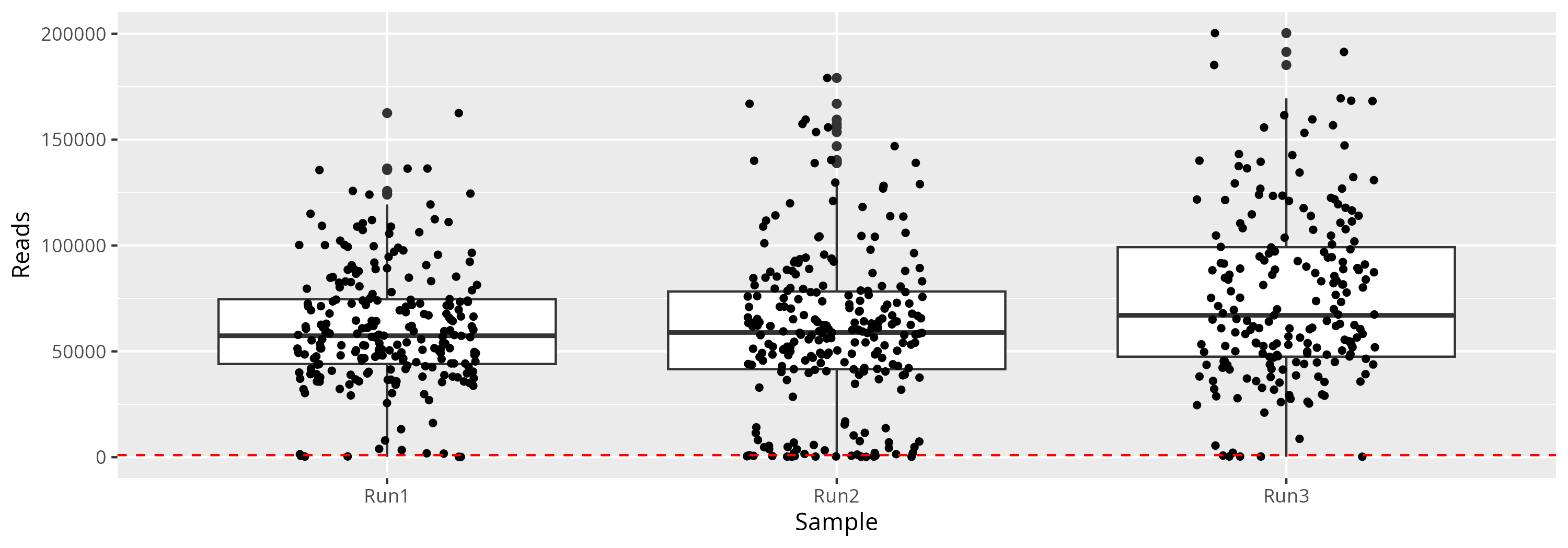
27 samples have less than 1,000 reads (Table 1)
raw_data %>% filter(num_seqs < 1000) %>% select(Sample, Run, num_seqs) %>% arrange(num_seqs) %>% kbl() %>% kable_styling(full_width = F)| Sample | Run | num_seqs |
|---|---|---|
| 19950J61 | Run1 | 157 |
| H2ObisRUN2 | Run2 | 194 |
| H2ORUN1 | Run1 | 203 |
| 18894rJ61 | Run2 | 214 |
| 18826J61 | Run2 | 229 |
| H2ObisRUN3 | Run3 | 235 |
| 18884J61 | Run2 | 238 |
| 18852J61 | Run2 | 301 |
| 19942J47 | Run2 | 301 |
| 19956J2 | Run1 | 311 |
| 18892J61 | Run2 | 322 |
| H2O4RUN3 | Run3 | 336 |
| H2ORUN3 | Run3 | 338 |
| H2OterRUN3 | Run3 | 348 |
| H2ObRUN1 | Run1 | 364 |
| 18900J61 | Run2 | 373 |
| 19938J47 | Run2 | 377 |
| 19918J47 | Run2 | 470 |
| H2ORUN2 | Run2 | 533 |
| 19954J2 | Run1 | 572 |
| 19906J61 | Run2 | 582 |
| 18852J47 | Run2 | 662 |
| 18840J61 | Run2 | 668 |
| 12771J47 | Run2 | 707 |
| 19974J47 | Run2 | 738 |
| Pq9Tneg | Run3 | 779 |
| 18832J61 | Run2 | 931 |
Quality control
cd /work_projet/eggtomeat/RUN1
mkdir FASTQC LOGS
for i in /save_projet/eggtomeat/RAW_DATA/RUN1/*.fastq.gz ; do echo "conda activate fastqc-0.11.9 && fastqc $i -o FASTQC && conda deactivate" >> fastqc.sh ; done
qarray -cwd -V -N fastqc -o LOGS -e LOGS fastqc.sh
cd /work_projet/eggtomeat/RUN2
mkdir FASTQC LOGS
for i in /save_projet/eggtomeat/RAW_DATA/RUN2/*.fastq.gz ; do echo "conda activate fastqc-0.11.9 && fastqc $i -o FASTQC && conda deactivate" >> fastqc.sh ; done
qarray -cwd -V -N fastqc -o LOGS -e LOGS fastqc.sh
cd /work_projet/eggtomeat/RUN3
mkdir FASTQC LOGS
for i in /save_projet/eggtomeat/RAW_DATA/RUN3/*.fastq.gz ; do echo "conda activate fastqc-0.11.9 && fastqc $i -o FASTQC && conda deactivate" >> fastqc.sh ; done
qarray -cwd -V -N fastqc -o LOGS -e LOGS fastqc.sh
qsub -cwd -V -N multiqc -o LOGS -e LOGS -b y "conda activate multiqc-1.11 && multiqc RUN1/FASTQC RUN2/FASTQC/ RUN3/FASTQC -o MULTIQC && conda deactivate"Quality control shows heterogeneous metrics between samples. Some samples are very poorly sequenced (controls but also samples of interest). The sequencing quality of some samples is also poor after 150 base pairs. There are still some N’s in a few reads, we also notice the presence of Illumina adapters, indicating very small fragments. All of these poor quality reads will be dicarded with bioinformatics.
Bioinformatics
A combination of dada2
flowchart TB
style FROGS stroke:#5f999d
style dada2 stroke:#5f999d
database[(Databank)]-->affiliation
fastq[[Fastq]]
biom[[BIOM]]
tsv[[TSV]]
fastq-->seqkit
seqkit---plotQualityProfiles
subgraph dada2
direction TB
plotQualityProfiles-->filterAndTrim1[filterAndTrim]-->filterAndTrim2[filterAndTrim]-->learnErrors-->dada-->mergePairs-->makeSequenceTable
end
filterAndTrim1-->cutadapt-->filterAndTrim2
makeSequenceTable-->rc
subgraph FROGS
direction TB
rc[remove chimera]-->filters-->affiliation
end
affiliation-->biom
affiliation--> tsv
cd /work_projet/eggtomeat/
cp /work_home/orue/GIT/dada2_frogs/dada2_FROGS.Rmd .
R -e "rmarkdown::render('dada2_FROGS.Rmd', params=list(
author='Olivier Rué',
reference='/db/outils/FROGS/assignation/silva_138.1_16S_pintail100/silva_138.1_16S_pintail100.fasta',
region='16S',
min_reads=1000,
expname='EGGTOMEAT_RUN1',
input_directory='/save_projet/eggtomeat/RAW_DATA/RUN1/',
forward_primer='ACGGRAGGCWGCAG',
reverse_primer='TACCAGGGTATCTAATCCT',
output_directory='/work_projet/eggtomeat/RUN1/out_dada2_FROGS/',
min_abundance=0.00005,
its=FALSE,
threads=24),
output_file = '/work_projet/eggtomeat/RUN1/out_dada2_FROGS/report_run1.html')"
R -e "rmarkdown::render('dada2_FROGS.Rmd', params=list(
author='Olivier Rué',
reference='/db/outils/FROGS/assignation/silva_138.1_16S_pintail100/silva_138.1_16S_pintail100.fasta',
region='16S',
min_reads=1000,
expname='EGGTOMEAT_RUN2',
input_directory='/save_projet/eggtomeat/RAW_DATA/RUN2/',
forward_primer='ACGGRAGGCWGCAG',
reverse_primer='TACCAGGGTATCTAATCCT',
output_directory='/work_projet/eggtomeat/RUN2/out_dada2_FROGS/',
min_abundance=0.00005,
its=FALSE,
threads=24),
output_file = '/work_projet/eggtomeat/RUN2/out_dada2_FROGS/report_run2.html')"
R -e "rmarkdown::render('dada2_FROGS.Rmd', params=list(
author='Olivier Rué',
reference='/db/outils/FROGS/assignation/silva_138.1_16S_pintail100/silva_138.1_16S_pintail100.fasta',
region='16S',
min_reads=1000,
expname='EGGTOMEAT_RUN3',
input_directory='/save_projet/eggtomeat/RAW_DATA/RUN3/',
forward_primer='ACGGRAGGCWGCAG',
reverse_primer='TACCAGGGTATCTAATCCT',
output_directory='/work_projet/eggtomeat/RUN3/out_dada2_FROGS/',
min_abundance=0.00005,
its=FALSE,
threads=24),
output_file = '/work_projet/eggtomeat/RUN3/out_dada2_FROGS/report_run3.html')"Biostatistics
if(!file.exists("html/physeq.rds")){
for(run in c("RUN1","RUN2","RUN3")){
biomfile <- paste0("html/",run,"/FROGS/affiliations.biom")
frogs <- import_frogs(biomfile, taxMethod = "blast")
metadata <- read.table("data/metadata.tsv", row.names = 1, header = TRUE, sep = "\t", stringsAsFactors = FALSE)
sample_data(frogs) <- metadata
saveRDS(frogs, paste0("html/",run,"/",run,".rds"))
}
}All samples are merged…
if(!file.exists("html/physeq.rds")){
physeq <- merge_phyloseq(readRDS("html/RUN1/RUN1.rds"),readRDS("html/RUN2/RUN2.rds"),readRDS("html/RUN3/RUN3.rds"))
saveRDS(physeq, "html/physeq.rds")
}else{
physeq <- readRDS("html/physeq.rds")
}physeqphyloseq-class experiment-level object
otu_table() OTU Table: [ 2256 taxa and 651 samples ]
sample_data() Sample Data: [ 651 samples by 9 sample variables ]
tax_table() Taxonomy Table: [ 2256 taxa by 7 taxonomic ranks ]… and samples with less than 1000 reads are removed
physeq_final <- subset_samples(physeq, sample_sums(physeq) > 1000)
saveRDS(physeq_final,"html/physeq_final.rds")The available variables describing the samples are
names(sample_data(physeq_final))[1] "QCbannks" "ClusterRaw" "ClusterPF" "run_number"
[5] "breeding_type" "chicken_sex" "chicken_ID" "breeding_time"
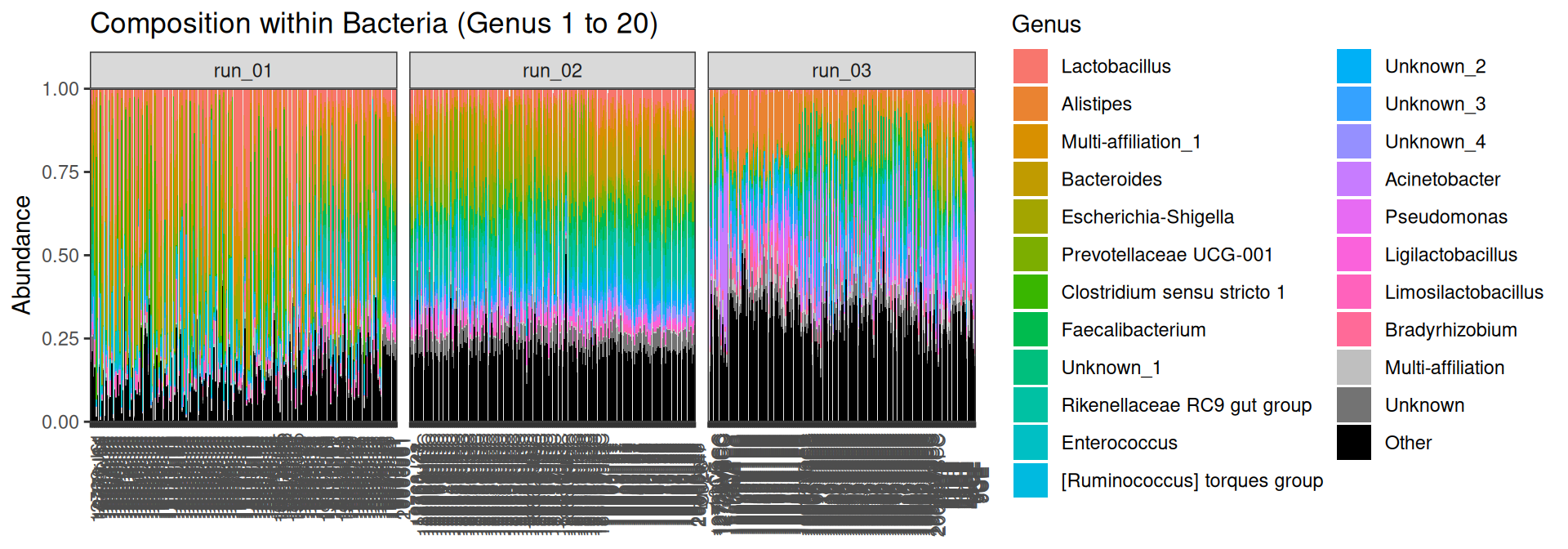
[9] "sample_type" p <- plot_composition(physeq = physeq_final, taxaRank1 = "Kingdom", taxaSet1 = "Bacteria", taxaRank2 = "Genus", numberOfTaxa = 20L, x = "Sample")Problematic taxa
taxa
TGGGGAATATTGCACAATGGGCGAAAGCCTGATGCAGCAACGCCGCGTGAGCGATGAAGGCCTTCGGGTCGTAAAGCTCTGTCCTCAAGGAAGATAATGACGGTACTTGAGGAGGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCTAGCGTTATCCGGAATTACTGGGCGTAAAGGGTGCGTAGGTGGTTTCTTAAGTCAGAGGTGAAAGGCTACGGCTCAACCGTAGTAAGCCTTTGAAACTGGGAAACTTGAGTGCAGGAGAGGAGAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTTGCGAAGGCGGCTCTCTGGACTGTAACTGACACTGAGGCACGAAAGCGTGGGGAGCAAAC TGGGGAATATTGCACAATGGGCGAAAGCCTGATGCAGCAACGCCGCGTGAGCGATGAAGGCCTTCGGGTCGTAAAGCTCTGTCCTCAAGGAAGATAATGACGGTACTTGAGGAGGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCTAGCGTTATCCGGAATTACTGGGCGTAAAGGGTGCGTAGGTGGTTTCTTAAGTCAGAGGTGAAAGGCTACGGCTCAACCGTAGTAAGCCTTTGAAACTGGGAAACTTGAGTGCAGGAGAGGAGAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTTGCGAAGGCGGCTCTCTGGACTGTAACTGACACTGAGGCACGAAAGCGTGGGGAGCAAAC
TCGGGAATATTGCGCAATGGAGGAAACTCTGACGCAGTGACGCCGCGTATAGGAAGAAGGTTTTCGGATTGTAAACTATTGTCGTTAGGGAAGATAAAAGACAGTACCTAAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTATCCGGATTTATTGGGTGTAAAGGGTGCGTAGACGGGATATTAAGTTAGTTGTGAAATCCCTCGGCTTAACTGAGGAACTGCAACTAAAACTGGTATTCTTGAGTGTTGGAGAGGAAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCGGTGGCGAAGGCGACTTTCTGGACAATAACTGACGTTGAGGCACGAAAGTGTGGGGAGCAAAC TCGGGAATATTGCGCAATGGAGGAAACTCTGACGCAGTGACGCCGCGTATAGGAAGAAGGTTTTCGGATTGTAAACTATTGTCGTTAGGGAAGATAAAAGACAGTACCTAAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTATCCGGATTTATTGGGTGTAAAGGGTGCGTAGACGGGATATTAAGTTAGTTGTGAAATCCCTCGGCTTAACTGAGGAACTGCAACTAAAACTGGTATTCTTGAGTGTTGGAGAGGAAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCGGTGGCGAAGGCGACTTTCTGGACAATAACTGACGTTGAGGCACGAAAGTGTGGGGAGCAAAC
TGGGGAATATTGGGCAATGGGCGAAAGCCTTACCCAGCAATGCCGCGTGAGTGAAGAAGGTCTTCGGATTGTAAAGCTCTTTGATTGGGGACGAGTAGAAGACGGTACCCAAGGAACAAGCCCCGGCTAACTATGTGCCAGCAGCCGCGGTAATACATAGGGGGCGAGCGTTGTCCGGAATGACTGGGCGTAAAGGGTGTGTAGGCGGTTTGGCAAGTTAGAAGTGTAATACCCAGGGCTTAACTCGGGTGCTGCTTCTAAAACTACCTGACTTGAGTGTCGGAGAGGAAAATGGAATTCCCAGTGTAGCGGTAGAATGCACAGATATTGGGAGGAACACCGGAGGCGAAAGCGATTTTCTGGACGACAACTGACGCTGAGGCACGAAAGCGTGGGGATCAAAC TGGGGAATATTGGGCAATGGGCGAAAGCCTTACCCAGCAATGCCGCGTGAGTGAAGAAGGTCTTCGGATTGTAAAGCTCTTTGATTGGGGACGAGTAGAAGACGGTACCCAAGGAACAAGCCCCGGCTAACTATGTGCCAGCAGCCGCGGTAATACATAGGGGGCGAGCGTTGTCCGGAATGACTGGGCGTAAAGGGTGTGTAGGCGGTTTGGCAAGTTAGAAGTGTAATACCCAGGGCTTAACTCGGGTGCTGCTTCTAAAACTACCTGACTTGAGTGTCGGAGAGGAAAATGGAATTCCCAGTGTAGCGGTAGAATGCACAGATATTGGGAGGAACACCGGAGGCGAAAGCGATTTTCTGGACGACAACTGACGCTGAGGCACGAAAGCGTGGGGATCAAAC
TGGGGAATATTGCACAATGGGGGAAACCCTGATGCAGCGACGCCGCGTGAGTGAAGAAGTATTTCGGTATGTAAAGCTCTATCAGCAGGGAAGAAAATGACGGTACCTGACTAAGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCAAGCGTTATCCGGATTTACTGGGTGTAAAGGGAGCGTAGACGGCGAAGCAAGTCTGAAGTGAAAACCCAGGGCTCAACCCTGGGACTGCTTTGGAAACTGTTTTGCTAGAGTGCTGGAGAGGTAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTGGCGAAGGCGGCTTACTGGACAGTAACTGACGTTGAGGCTCGAAAGCGTGGGGAGCAAAC TGGGGAATATTGCACAATGGGGGAAACCCTGATGCAGCGACGCCGCGTGAGTGAAGAAGTATTTCGGTATGTAAAGCTCTATCAGCAGGGAAGAAAATGACGGTACCTGACTAAGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCAAGCGTTATCCGGATTTACTGGGTGTAAAGGGAGCGTAGACGGCGAAGCAAGTCTGAAGTGAAAACCCAGGGCTCAACCCTGGGACTGCTTTGGAAACTGTTTTGCTAGAGTGCTGGAGAGGTAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTGGCGAAGGCGGCTTACTGGACAGTAACTGACGTTGAGGCTCGAAAGCGTGGGGAGCAAAC
TGAGGGATATTGGTCAATGGGGGAAACCCTGAACCAGCAACGCCGCGTGAGGGATGACGGCCTTCGGGTTGTAAACCTCTGTCCTCTGTGAAGATAATGACGGTAGCAGAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTGTCCGGATTTACTGGGTGTAAAGGGTGCGTAGGCGGTTTGGTAAGTCAGAAGTGAAATCCATGGGCTTAACCCATGAACTGCTTTTGAAACTATCGAACTTGAGTGAAGTAGAGGTAGGCGGAATTCCCGGTGTAGCGGTGAAATGCGTAGAGATCGGGAGGAACACCAGTGGCGAAGGCGGCCTACTGGGCTTTAACTGACGCTGAGGCACGAAAGCATGGGTAGCAAAC TGAGGGATATTGGTCAATGGGGGAAACCCTGAACCAGCAACGCCGCGTGAGGGATGACGGCCTTCGGGTTGTAAACCTCTGTCCTCTGTGAAGATAATGACGGTAGCAGAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTGTCCGGATTTACTGGGTGTAAAGGGTGCGTAGGCGGTTTGGTAAGTCAGAAGTGAAATCCATGGGCTTAACCCATGAACTGCTTTTGAAACTATCGAACTTGAGTGAAGTAGAGGTAGGCGGAATTCCCGGTGTAGCGGTGAAATGCGTAGAGATCGGGAGGAACACCAGTGGCGAAGGCGGCCTACTGGGCTTTAACTGACGCTGAGGCACGAAAGCATGGGTAGCAAAC
Kingdom
TGGGGAATATTGCACAATGGGCGAAAGCCTGATGCAGCAACGCCGCGTGAGCGATGAAGGCCTTCGGGTCGTAAAGCTCTGTCCTCAAGGAAGATAATGACGGTACTTGAGGAGGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCTAGCGTTATCCGGAATTACTGGGCGTAAAGGGTGCGTAGGTGGTTTCTTAAGTCAGAGGTGAAAGGCTACGGCTCAACCGTAGTAAGCCTTTGAAACTGGGAAACTTGAGTGCAGGAGAGGAGAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTTGCGAAGGCGGCTCTCTGGACTGTAACTGACACTGAGGCACGAAAGCGTGGGGAGCAAAC Bacteria
TCGGGAATATTGCGCAATGGAGGAAACTCTGACGCAGTGACGCCGCGTATAGGAAGAAGGTTTTCGGATTGTAAACTATTGTCGTTAGGGAAGATAAAAGACAGTACCTAAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTATCCGGATTTATTGGGTGTAAAGGGTGCGTAGACGGGATATTAAGTTAGTTGTGAAATCCCTCGGCTTAACTGAGGAACTGCAACTAAAACTGGTATTCTTGAGTGTTGGAGAGGAAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCGGTGGCGAAGGCGACTTTCTGGACAATAACTGACGTTGAGGCACGAAAGTGTGGGGAGCAAAC Bacteria
TGGGGAATATTGGGCAATGGGCGAAAGCCTTACCCAGCAATGCCGCGTGAGTGAAGAAGGTCTTCGGATTGTAAAGCTCTTTGATTGGGGACGAGTAGAAGACGGTACCCAAGGAACAAGCCCCGGCTAACTATGTGCCAGCAGCCGCGGTAATACATAGGGGGCGAGCGTTGTCCGGAATGACTGGGCGTAAAGGGTGTGTAGGCGGTTTGGCAAGTTAGAAGTGTAATACCCAGGGCTTAACTCGGGTGCTGCTTCTAAAACTACCTGACTTGAGTGTCGGAGAGGAAAATGGAATTCCCAGTGTAGCGGTAGAATGCACAGATATTGGGAGGAACACCGGAGGCGAAAGCGATTTTCTGGACGACAACTGACGCTGAGGCACGAAAGCGTGGGGATCAAAC Bacteria
TGGGGAATATTGCACAATGGGGGAAACCCTGATGCAGCGACGCCGCGTGAGTGAAGAAGTATTTCGGTATGTAAAGCTCTATCAGCAGGGAAGAAAATGACGGTACCTGACTAAGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCAAGCGTTATCCGGATTTACTGGGTGTAAAGGGAGCGTAGACGGCGAAGCAAGTCTGAAGTGAAAACCCAGGGCTCAACCCTGGGACTGCTTTGGAAACTGTTTTGCTAGAGTGCTGGAGAGGTAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTGGCGAAGGCGGCTTACTGGACAGTAACTGACGTTGAGGCTCGAAAGCGTGGGGAGCAAAC Bacteria
TGAGGGATATTGGTCAATGGGGGAAACCCTGAACCAGCAACGCCGCGTGAGGGATGACGGCCTTCGGGTTGTAAACCTCTGTCCTCTGTGAAGATAATGACGGTAGCAGAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTGTCCGGATTTACTGGGTGTAAAGGGTGCGTAGGCGGTTTGGTAAGTCAGAAGTGAAATCCATGGGCTTAACCCATGAACTGCTTTTGAAACTATCGAACTTGAGTGAAGTAGAGGTAGGCGGAATTCCCGGTGTAGCGGTGAAATGCGTAGAGATCGGGAGGAACACCAGTGGCGAAGGCGGCCTACTGGGCTTTAACTGACGCTGAGGCACGAAAGCATGGGTAGCAAAC Bacteria
Phylum
TGGGGAATATTGCACAATGGGCGAAAGCCTGATGCAGCAACGCCGCGTGAGCGATGAAGGCCTTCGGGTCGTAAAGCTCTGTCCTCAAGGAAGATAATGACGGTACTTGAGGAGGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCTAGCGTTATCCGGAATTACTGGGCGTAAAGGGTGCGTAGGTGGTTTCTTAAGTCAGAGGTGAAAGGCTACGGCTCAACCGTAGTAAGCCTTTGAAACTGGGAAACTTGAGTGCAGGAGAGGAGAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTTGCGAAGGCGGCTCTCTGGACTGTAACTGACACTGAGGCACGAAAGCGTGGGGAGCAAAC Firmicutes
TCGGGAATATTGCGCAATGGAGGAAACTCTGACGCAGTGACGCCGCGTATAGGAAGAAGGTTTTCGGATTGTAAACTATTGTCGTTAGGGAAGATAAAAGACAGTACCTAAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTATCCGGATTTATTGGGTGTAAAGGGTGCGTAGACGGGATATTAAGTTAGTTGTGAAATCCCTCGGCTTAACTGAGGAACTGCAACTAAAACTGGTATTCTTGAGTGTTGGAGAGGAAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCGGTGGCGAAGGCGACTTTCTGGACAATAACTGACGTTGAGGCACGAAAGTGTGGGGAGCAAAC Firmicutes
TGGGGAATATTGGGCAATGGGCGAAAGCCTTACCCAGCAATGCCGCGTGAGTGAAGAAGGTCTTCGGATTGTAAAGCTCTTTGATTGGGGACGAGTAGAAGACGGTACCCAAGGAACAAGCCCCGGCTAACTATGTGCCAGCAGCCGCGGTAATACATAGGGGGCGAGCGTTGTCCGGAATGACTGGGCGTAAAGGGTGTGTAGGCGGTTTGGCAAGTTAGAAGTGTAATACCCAGGGCTTAACTCGGGTGCTGCTTCTAAAACTACCTGACTTGAGTGTCGGAGAGGAAAATGGAATTCCCAGTGTAGCGGTAGAATGCACAGATATTGGGAGGAACACCGGAGGCGAAAGCGATTTTCTGGACGACAACTGACGCTGAGGCACGAAAGCGTGGGGATCAAAC Firmicutes
TGGGGAATATTGCACAATGGGGGAAACCCTGATGCAGCGACGCCGCGTGAGTGAAGAAGTATTTCGGTATGTAAAGCTCTATCAGCAGGGAAGAAAATGACGGTACCTGACTAAGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCAAGCGTTATCCGGATTTACTGGGTGTAAAGGGAGCGTAGACGGCGAAGCAAGTCTGAAGTGAAAACCCAGGGCTCAACCCTGGGACTGCTTTGGAAACTGTTTTGCTAGAGTGCTGGAGAGGTAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTGGCGAAGGCGGCTTACTGGACAGTAACTGACGTTGAGGCTCGAAAGCGTGGGGAGCAAAC Firmicutes
TGAGGGATATTGGTCAATGGGGGAAACCCTGAACCAGCAACGCCGCGTGAGGGATGACGGCCTTCGGGTTGTAAACCTCTGTCCTCTGTGAAGATAATGACGGTAGCAGAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTGTCCGGATTTACTGGGTGTAAAGGGTGCGTAGGCGGTTTGGTAAGTCAGAAGTGAAATCCATGGGCTTAACCCATGAACTGCTTTTGAAACTATCGAACTTGAGTGAAGTAGAGGTAGGCGGAATTCCCGGTGTAGCGGTGAAATGCGTAGAGATCGGGAGGAACACCAGTGGCGAAGGCGGCCTACTGGGCTTTAACTGACGCTGAGGCACGAAAGCATGGGTAGCAAAC Firmicutes
Class
TGGGGAATATTGCACAATGGGCGAAAGCCTGATGCAGCAACGCCGCGTGAGCGATGAAGGCCTTCGGGTCGTAAAGCTCTGTCCTCAAGGAAGATAATGACGGTACTTGAGGAGGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCTAGCGTTATCCGGAATTACTGGGCGTAAAGGGTGCGTAGGTGGTTTCTTAAGTCAGAGGTGAAAGGCTACGGCTCAACCGTAGTAAGCCTTTGAAACTGGGAAACTTGAGTGCAGGAGAGGAGAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTTGCGAAGGCGGCTCTCTGGACTGTAACTGACACTGAGGCACGAAAGCGTGGGGAGCAAAC Clostridia
TCGGGAATATTGCGCAATGGAGGAAACTCTGACGCAGTGACGCCGCGTATAGGAAGAAGGTTTTCGGATTGTAAACTATTGTCGTTAGGGAAGATAAAAGACAGTACCTAAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTATCCGGATTTATTGGGTGTAAAGGGTGCGTAGACGGGATATTAAGTTAGTTGTGAAATCCCTCGGCTTAACTGAGGAACTGCAACTAAAACTGGTATTCTTGAGTGTTGGAGAGGAAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCGGTGGCGAAGGCGACTTTCTGGACAATAACTGACGTTGAGGCACGAAAGTGTGGGGAGCAAAC Clostridia
TGGGGAATATTGGGCAATGGGCGAAAGCCTTACCCAGCAATGCCGCGTGAGTGAAGAAGGTCTTCGGATTGTAAAGCTCTTTGATTGGGGACGAGTAGAAGACGGTACCCAAGGAACAAGCCCCGGCTAACTATGTGCCAGCAGCCGCGGTAATACATAGGGGGCGAGCGTTGTCCGGAATGACTGGGCGTAAAGGGTGTGTAGGCGGTTTGGCAAGTTAGAAGTGTAATACCCAGGGCTTAACTCGGGTGCTGCTTCTAAAACTACCTGACTTGAGTGTCGGAGAGGAAAATGGAATTCCCAGTGTAGCGGTAGAATGCACAGATATTGGGAGGAACACCGGAGGCGAAAGCGATTTTCTGGACGACAACTGACGCTGAGGCACGAAAGCGTGGGGATCAAAC Clostridia
TGGGGAATATTGCACAATGGGGGAAACCCTGATGCAGCGACGCCGCGTGAGTGAAGAAGTATTTCGGTATGTAAAGCTCTATCAGCAGGGAAGAAAATGACGGTACCTGACTAAGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCAAGCGTTATCCGGATTTACTGGGTGTAAAGGGAGCGTAGACGGCGAAGCAAGTCTGAAGTGAAAACCCAGGGCTCAACCCTGGGACTGCTTTGGAAACTGTTTTGCTAGAGTGCTGGAGAGGTAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTGGCGAAGGCGGCTTACTGGACAGTAACTGACGTTGAGGCTCGAAAGCGTGGGGAGCAAAC Clostridia
TGAGGGATATTGGTCAATGGGGGAAACCCTGAACCAGCAACGCCGCGTGAGGGATGACGGCCTTCGGGTTGTAAACCTCTGTCCTCTGTGAAGATAATGACGGTAGCAGAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTGTCCGGATTTACTGGGTGTAAAGGGTGCGTAGGCGGTTTGGTAAGTCAGAAGTGAAATCCATGGGCTTAACCCATGAACTGCTTTTGAAACTATCGAACTTGAGTGAAGTAGAGGTAGGCGGAATTCCCGGTGTAGCGGTGAAATGCGTAGAGATCGGGAGGAACACCAGTGGCGAAGGCGGCCTACTGGGCTTTAACTGACGCTGAGGCACGAAAGCATGGGTAGCAAAC Clostridia
Order
TGGGGAATATTGCACAATGGGCGAAAGCCTGATGCAGCAACGCCGCGTGAGCGATGAAGGCCTTCGGGTCGTAAAGCTCTGTCCTCAAGGAAGATAATGACGGTACTTGAGGAGGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCTAGCGTTATCCGGAATTACTGGGCGTAAAGGGTGCGTAGGTGGTTTCTTAAGTCAGAGGTGAAAGGCTACGGCTCAACCGTAGTAAGCCTTTGAAACTGGGAAACTTGAGTGCAGGAGAGGAGAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTTGCGAAGGCGGCTCTCTGGACTGTAACTGACACTGAGGCACGAAAGCGTGGGGAGCAAAC Peptostreptococcales-Tissierellales
TCGGGAATATTGCGCAATGGAGGAAACTCTGACGCAGTGACGCCGCGTATAGGAAGAAGGTTTTCGGATTGTAAACTATTGTCGTTAGGGAAGATAAAAGACAGTACCTAAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTATCCGGATTTATTGGGTGTAAAGGGTGCGTAGACGGGATATTAAGTTAGTTGTGAAATCCCTCGGCTTAACTGAGGAACTGCAACTAAAACTGGTATTCTTGAGTGTTGGAGAGGAAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCGGTGGCGAAGGCGACTTTCTGGACAATAACTGACGTTGAGGCACGAAAGTGTGGGGAGCAAAC Clostridia UCG-014
TGGGGAATATTGGGCAATGGGCGAAAGCCTTACCCAGCAATGCCGCGTGAGTGAAGAAGGTCTTCGGATTGTAAAGCTCTTTGATTGGGGACGAGTAGAAGACGGTACCCAAGGAACAAGCCCCGGCTAACTATGTGCCAGCAGCCGCGGTAATACATAGGGGGCGAGCGTTGTCCGGAATGACTGGGCGTAAAGGGTGTGTAGGCGGTTTGGCAAGTTAGAAGTGTAATACCCAGGGCTTAACTCGGGTGCTGCTTCTAAAACTACCTGACTTGAGTGTCGGAGAGGAAAATGGAATTCCCAGTGTAGCGGTAGAATGCACAGATATTGGGAGGAACACCGGAGGCGAAAGCGATTTTCTGGACGACAACTGACGCTGAGGCACGAAAGCGTGGGGATCAAAC Clostridia vadinBB60 group
TGGGGAATATTGCACAATGGGGGAAACCCTGATGCAGCGACGCCGCGTGAGTGAAGAAGTATTTCGGTATGTAAAGCTCTATCAGCAGGGAAGAAAATGACGGTACCTGACTAAGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCAAGCGTTATCCGGATTTACTGGGTGTAAAGGGAGCGTAGACGGCGAAGCAAGTCTGAAGTGAAAACCCAGGGCTCAACCCTGGGACTGCTTTGGAAACTGTTTTGCTAGAGTGCTGGAGAGGTAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTGGCGAAGGCGGCTTACTGGACAGTAACTGACGTTGAGGCTCGAAAGCGTGGGGAGCAAAC Lachnospirales
TGAGGGATATTGGTCAATGGGGGAAACCCTGAACCAGCAACGCCGCGTGAGGGATGACGGCCTTCGGGTTGTAAACCTCTGTCCTCTGTGAAGATAATGACGGTAGCAGAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTGTCCGGATTTACTGGGTGTAAAGGGTGCGTAGGCGGTTTGGTAAGTCAGAAGTGAAATCCATGGGCTTAACCCATGAACTGCTTTTGAAACTATCGAACTTGAGTGAAGTAGAGGTAGGCGGAATTCCCGGTGTAGCGGTGAAATGCGTAGAGATCGGGAGGAACACCAGTGGCGAAGGCGGCCTACTGGGCTTTAACTGACGCTGAGGCACGAAAGCATGGGTAGCAAAC Oscillospirales
Family
TGGGGAATATTGCACAATGGGCGAAAGCCTGATGCAGCAACGCCGCGTGAGCGATGAAGGCCTTCGGGTCGTAAAGCTCTGTCCTCAAGGAAGATAATGACGGTACTTGAGGAGGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCTAGCGTTATCCGGAATTACTGGGCGTAAAGGGTGCGTAGGTGGTTTCTTAAGTCAGAGGTGAAAGGCTACGGCTCAACCGTAGTAAGCCTTTGAAACTGGGAAACTTGAGTGCAGGAGAGGAGAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTTGCGAAGGCGGCTCTCTGGACTGTAACTGACACTGAGGCACGAAAGCGTGGGGAGCAAAC Peptostreptococcaceae
TCGGGAATATTGCGCAATGGAGGAAACTCTGACGCAGTGACGCCGCGTATAGGAAGAAGGTTTTCGGATTGTAAACTATTGTCGTTAGGGAAGATAAAAGACAGTACCTAAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTATCCGGATTTATTGGGTGTAAAGGGTGCGTAGACGGGATATTAAGTTAGTTGTGAAATCCCTCGGCTTAACTGAGGAACTGCAACTAAAACTGGTATTCTTGAGTGTTGGAGAGGAAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCGGTGGCGAAGGCGACTTTCTGGACAATAACTGACGTTGAGGCACGAAAGTGTGGGGAGCAAAC Unknown
TGGGGAATATTGGGCAATGGGCGAAAGCCTTACCCAGCAATGCCGCGTGAGTGAAGAAGGTCTTCGGATTGTAAAGCTCTTTGATTGGGGACGAGTAGAAGACGGTACCCAAGGAACAAGCCCCGGCTAACTATGTGCCAGCAGCCGCGGTAATACATAGGGGGCGAGCGTTGTCCGGAATGACTGGGCGTAAAGGGTGTGTAGGCGGTTTGGCAAGTTAGAAGTGTAATACCCAGGGCTTAACTCGGGTGCTGCTTCTAAAACTACCTGACTTGAGTGTCGGAGAGGAAAATGGAATTCCCAGTGTAGCGGTAGAATGCACAGATATTGGGAGGAACACCGGAGGCGAAAGCGATTTTCTGGACGACAACTGACGCTGAGGCACGAAAGCGTGGGGATCAAAC Unknown
TGGGGAATATTGCACAATGGGGGAAACCCTGATGCAGCGACGCCGCGTGAGTGAAGAAGTATTTCGGTATGTAAAGCTCTATCAGCAGGGAAGAAAATGACGGTACCTGACTAAGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCAAGCGTTATCCGGATTTACTGGGTGTAAAGGGAGCGTAGACGGCGAAGCAAGTCTGAAGTGAAAACCCAGGGCTCAACCCTGGGACTGCTTTGGAAACTGTTTTGCTAGAGTGCTGGAGAGGTAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTGGCGAAGGCGGCTTACTGGACAGTAACTGACGTTGAGGCTCGAAAGCGTGGGGAGCAAAC Lachnospiraceae
TGAGGGATATTGGTCAATGGGGGAAACCCTGAACCAGCAACGCCGCGTGAGGGATGACGGCCTTCGGGTTGTAAACCTCTGTCCTCTGTGAAGATAATGACGGTAGCAGAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTGTCCGGATTTACTGGGTGTAAAGGGTGCGTAGGCGGTTTGGTAAGTCAGAAGTGAAATCCATGGGCTTAACCCATGAACTGCTTTTGAAACTATCGAACTTGAGTGAAGTAGAGGTAGGCGGAATTCCCGGTGTAGCGGTGAAATGCGTAGAGATCGGGAGGAACACCAGTGGCGAAGGCGGCCTACTGGGCTTTAACTGACGCTGAGGCACGAAAGCATGGGTAGCAAAC Ruminococcaceae
Genus
TGGGGAATATTGCACAATGGGCGAAAGCCTGATGCAGCAACGCCGCGTGAGCGATGAAGGCCTTCGGGTCGTAAAGCTCTGTCCTCAAGGAAGATAATGACGGTACTTGAGGAGGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCTAGCGTTATCCGGAATTACTGGGCGTAAAGGGTGCGTAGGTGGTTTCTTAAGTCAGAGGTGAAAGGCTACGGCTCAACCGTAGTAAGCCTTTGAAACTGGGAAACTTGAGTGCAGGAGAGGAGAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTTGCGAAGGCGGCTCTCTGGACTGTAACTGACACTGAGGCACGAAAGCGTGGGGAGCAAAC Multi-affiliation
TCGGGAATATTGCGCAATGGAGGAAACTCTGACGCAGTGACGCCGCGTATAGGAAGAAGGTTTTCGGATTGTAAACTATTGTCGTTAGGGAAGATAAAAGACAGTACCTAAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTATCCGGATTTATTGGGTGTAAAGGGTGCGTAGACGGGATATTAAGTTAGTTGTGAAATCCCTCGGCTTAACTGAGGAACTGCAACTAAAACTGGTATTCTTGAGTGTTGGAGAGGAAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCGGTGGCGAAGGCGACTTTCTGGACAATAACTGACGTTGAGGCACGAAAGTGTGGGGAGCAAAC Unknown
TGGGGAATATTGGGCAATGGGCGAAAGCCTTACCCAGCAATGCCGCGTGAGTGAAGAAGGTCTTCGGATTGTAAAGCTCTTTGATTGGGGACGAGTAGAAGACGGTACCCAAGGAACAAGCCCCGGCTAACTATGTGCCAGCAGCCGCGGTAATACATAGGGGGCGAGCGTTGTCCGGAATGACTGGGCGTAAAGGGTGTGTAGGCGGTTTGGCAAGTTAGAAGTGTAATACCCAGGGCTTAACTCGGGTGCTGCTTCTAAAACTACCTGACTTGAGTGTCGGAGAGGAAAATGGAATTCCCAGTGTAGCGGTAGAATGCACAGATATTGGGAGGAACACCGGAGGCGAAAGCGATTTTCTGGACGACAACTGACGCTGAGGCACGAAAGCGTGGGGATCAAAC Unknown
TGGGGAATATTGCACAATGGGGGAAACCCTGATGCAGCGACGCCGCGTGAGTGAAGAAGTATTTCGGTATGTAAAGCTCTATCAGCAGGGAAGAAAATGACGGTACCTGACTAAGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCAAGCGTTATCCGGATTTACTGGGTGTAAAGGGAGCGTAGACGGCGAAGCAAGTCTGAAGTGAAAACCCAGGGCTCAACCCTGGGACTGCTTTGGAAACTGTTTTGCTAGAGTGCTGGAGAGGTAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTGGCGAAGGCGGCTTACTGGACAGTAACTGACGTTGAGGCTCGAAAGCGTGGGGAGCAAAC Unknown
TGAGGGATATTGGTCAATGGGGGAAACCCTGAACCAGCAACGCCGCGTGAGGGATGACGGCCTTCGGGTTGTAAACCTCTGTCCTCTGTGAAGATAATGACGGTAGCAGAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTGTCCGGATTTACTGGGTGTAAAGGGTGCGTAGGCGGTTTGGTAAGTCAGAAGTGAAATCCATGGGCTTAACCCATGAACTGCTTTTGAAACTATCGAACTTGAGTGAAGTAGAGGTAGGCGGAATTCCCGGTGTAGCGGTGAAATGCGTAGAGATCGGGAGGAACACCAGTGGCGAAGGCGGCCTACTGGGCTTTAACTGACGCTGAGGCACGAAAGCATGGGTAGCAAAC Unknown
rank
TGGGGAATATTGCACAATGGGCGAAAGCCTGATGCAGCAACGCCGCGTGAGCGATGAAGGCCTTCGGGTCGTAAAGCTCTGTCCTCAAGGAAGATAATGACGGTACTTGAGGAGGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCTAGCGTTATCCGGAATTACTGGGCGTAAAGGGTGCGTAGGTGGTTTCTTAAGTCAGAGGTGAAAGGCTACGGCTCAACCGTAGTAAGCCTTTGAAACTGGGAAACTTGAGTGCAGGAGAGGAGAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTTGCGAAGGCGGCTCTCTGGACTGTAACTGACACTGAGGCACGAAAGCGTGGGGAGCAAAC 3
TCGGGAATATTGCGCAATGGAGGAAACTCTGACGCAGTGACGCCGCGTATAGGAAGAAGGTTTTCGGATTGTAAACTATTGTCGTTAGGGAAGATAAAAGACAGTACCTAAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTATCCGGATTTATTGGGTGTAAAGGGTGCGTAGACGGGATATTAAGTTAGTTGTGAAATCCCTCGGCTTAACTGAGGAACTGCAACTAAAACTGGTATTCTTGAGTGTTGGAGAGGAAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCGGTGGCGAAGGCGACTTTCTGGACAATAACTGACGTTGAGGCACGAAAGTGTGGGGAGCAAAC 9
TGGGGAATATTGGGCAATGGGCGAAAGCCTTACCCAGCAATGCCGCGTGAGTGAAGAAGGTCTTCGGATTGTAAAGCTCTTTGATTGGGGACGAGTAGAAGACGGTACCCAAGGAACAAGCCCCGGCTAACTATGTGCCAGCAGCCGCGGTAATACATAGGGGGCGAGCGTTGTCCGGAATGACTGGGCGTAAAGGGTGTGTAGGCGGTTTGGCAAGTTAGAAGTGTAATACCCAGGGCTTAACTCGGGTGCTGCTTCTAAAACTACCTGACTTGAGTGTCGGAGAGGAAAATGGAATTCCCAGTGTAGCGGTAGAATGCACAGATATTGGGAGGAACACCGGAGGCGAAAGCGATTTTCTGGACGACAACTGACGCTGAGGCACGAAAGCGTGGGGATCAAAC 13
TGGGGAATATTGCACAATGGGGGAAACCCTGATGCAGCGACGCCGCGTGAGTGAAGAAGTATTTCGGTATGTAAAGCTCTATCAGCAGGGAAGAAAATGACGGTACCTGACTAAGAAGCCCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGGGCAAGCGTTATCCGGATTTACTGGGTGTAAAGGGAGCGTAGACGGCGAAGCAAGTCTGAAGTGAAAACCCAGGGCTCAACCCTGGGACTGCTTTGGAAACTGTTTTGCTAGAGTGCTGGAGAGGTAAGTGGAATTCCTAGTGTAGCGGTGAAATGCGTAGATATTAGGAGGAACACCAGTGGCGAAGGCGGCTTACTGGACAGTAACTGACGTTGAGGCTCGAAAGCGTGGGGAGCAAAC 14
TGAGGGATATTGGTCAATGGGGGAAACCCTGAACCAGCAACGCCGCGTGAGGGATGACGGCCTTCGGGTTGTAAACCTCTGTCCTCTGTGAAGATAATGACGGTAGCAGAGGAGGAAGCTCCGGCTAACTACGTGCCAGCAGCCGCGGTAATACGTAGGGAGCAAGCGTTGTCCGGATTTACTGGGTGTAAAGGGTGCGTAGGCGGTTTGGTAAGTCAGAAGTGAAATCCATGGGCTTAACCCATGAACTGCTTTTGAAACTATCGAACTTGAGTGAAGTAGAGGTAGGCGGAATTCCCGGTGTAGCGGTGAAATGCGTAGAGATCGGGAGGAACACCAGTGGCGAAGGCGGCCTACTGGGCTTTAACTGACGCTGAGGCACGAAAGCATGGGTAGCAAAC 15p + facet_grid(". ~ run_number", scales = "free_x", space = "free")