Show the code
#Use packages and functions
library(tidyverse)
library(edgeR)
library(ggplot2)
library(kableExtra)
library(ExperimentSubset)
library(colorspace)
library(openxlsx) # manipulation excel files
#sessionInfo()Experiment with BC
Migale bioinformatics facility
Migale bioinformatics facility
May 2, 2023
February 19, 2025
We used datasets created at the exploration and normalisation step from BC experiment. Genes with too low expression were filtered. A normalization factor is calculated to take into account the different sizes of the sequencing banks (i.e. the total read count) and the distribution of reads per sample on sequencing run, as discussed [1]. Read counts were normalized using trimmed mean of M-values (TMM) method [2] from edgeR
BC_G_R1 BC_G_R2 BC_G_R3 BC_R_R1 BC_R_R2 BC_R_R3
gene-BBCT_RS00005 2244 1589 2602 1720 1491 1500
gene-BBCT_RS00010 3481 2690 3314 944 883 786
gene-BBCT_RS00015 293 224 348 86 91 60Above an extract from the expression matrix which contains expression on 1693 expressed genes and 6 samples from BC experiment.
The aim is to identify the genes differentially expressed between treatment and Glucose.
The differential analysis is based on a Generalized Linear Model (GLM) to take into account the distribution of count-type data (non-normality of the data) and over-dispersion. The regression coefficients of the factors are estimated by a log-linear model.
Using edgeR
dge$samples$group <- as.factor(dge$samples$group)
dge$samples$group <- relevel(dge$samples$group, ref = paste(params$my.interest, "G", sep="_"))
# levels(dge$samples$group)
#matrix design: one factor group
design <- model.matrix(~ 0 + group, data = dge$samples)
colnames(design) <- gsub(paste0("group", params$my.interest, "_"), "", colnames(design))
# designWe set robust=TRUE when estimating the dispersion. This has no effect on the downstream analysis, but is nevertheless very useful for identifying outliers from the trend of the mean dispersion of the negative binomial distribution.
We used GLM model [3] to perform likelihood ratio tests for each contrast defined as treatment versus Glucose control treatment. List of differentially expressed genes were obtained after controlling the false discovery rate with a Benjamini-Hochberg correction [4] at 0.05 threshold.
Histogram of raw p-values and mean-difference plot (also called MA-plot) are useful graphs to validate the model.
#only the trended dispersion is used under the QL
method <- "LRT"
#fit <- glmFit(dge, dge$design)
# with specific design
fit <- glmFit(dge, design = design)
#head(fit$coefficients)
#head(fit$dispersion)
res<-list()
nDiffTotal <- NULL
#fit model for each contrast
for (ctr in 1:length(my.contrasts)){
qlf<- glmLRT(fit, contrast = my.contrasts[[ctr]])
topTags(qlf)
# Results, Nb of DE
#cat("contrast= ",names(my.contrasts)[ctr],"\n")
#print(summary(decideTestsDGE(qlf)))
nDiffTotal <- cbind(nDiffTotal, summary(decideTestsDGE(qlf)))
# results of tests
res[[ctr]]<-topTags(qlf,n=nrow(dge$counts),adjust.method="BH",sort.by="none")$table
names(res)[ctr]<-names(my.contrasts)[ctr]
# histogram of raw pvalues
my.title <- paste(names(my.contrasts)[ctr], "Up =", nDiffTotal[,ctr]["Up"], "Down =", nDiffTotal[,ctr]["Down"])
p <- ggplot(data = qlf$table, aes(x = PValue))
p <- p + geom_histogram(color="darkblue", fill="lightblue")
p <- p + theme_bw() + xlab("Raw pvalue")
p <- p + ggtitle(my.title)
print(p)
# MA plot
plotMD(qlf, main = my.title)
}
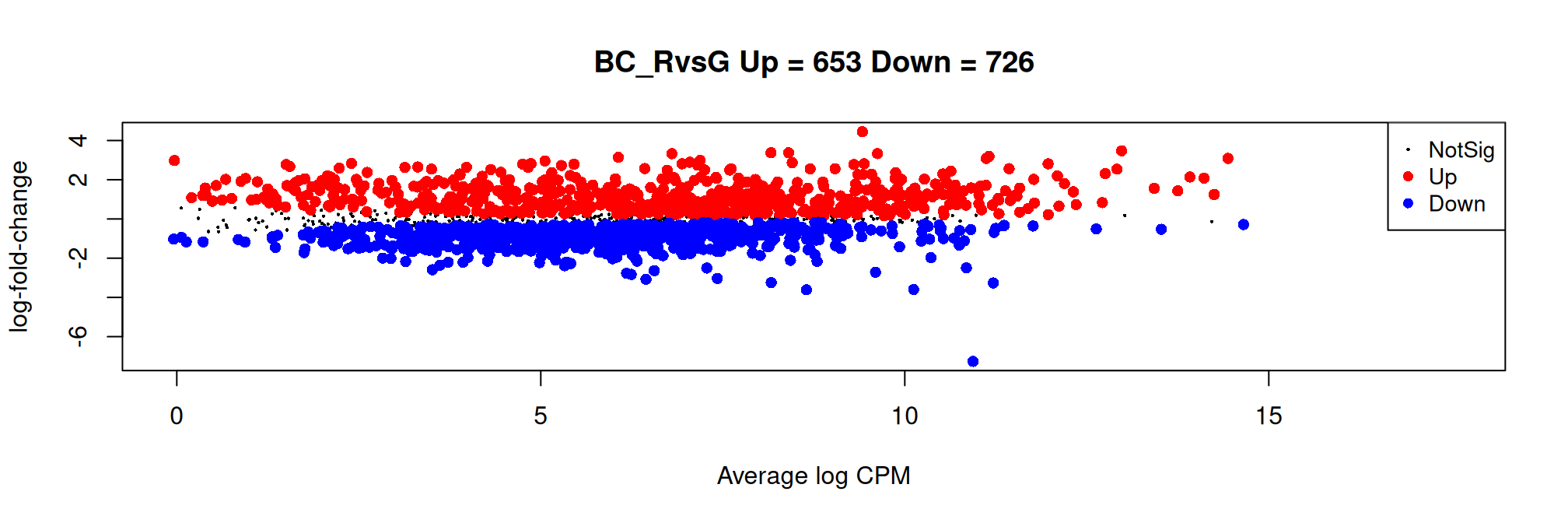
Number of features down/up and total:| BC_RvsG | |
|---|---|
| Down | 726 |
| NotSig | 314 |
| Up | 653 |
─ Session info ───────────────────────────────────────────────────────────────
setting value
version R version 4.4.2 (2024-10-31)
os Ubuntu 24.04.1 LTS
system x86_64, linux-gnu
ui X11
language (EN)
collate fr_FR.UTF-8
ctype fr_FR.UTF-8
tz Europe/Paris
date 2025-02-16
pandoc 3.1.1 @ /usr/lib/rstudio/resources/app/bin/quarto/bin/tools/ (via rmarkdown)
─ Packages ───────────────────────────────────────────────────────────────────
package * version date (UTC) lib source
Biobase * 2.64.0 2024-04-30 [1] Bioconductor 3.19 (R 4.4.0)
BiocGenerics * 0.50.0 2024-04-30 [1] Bioconductor 3.19 (R 4.4.0)
Biostrings * 2.72.1 2024-06-02 [1] Bioconductor 3.19 (R 4.4.0)
colorspace * 2.1-1 2024-07-26 [1] CRAN (R 4.4.0)
dplyr * 1.1.4 2023-11-17 [1] CRAN (R 4.4.0)
edgeR * 4.2.2 2024-10-13 [1] Bioconductor 3.19 (R 4.4.1)
ExperimentSubset * 1.14.0 2024-04-30 [1] Bioconductor 3.19 (R 4.4.2)
forcats * 1.0.0 2023-01-29 [1] CRAN (R 4.4.0)
GenomeInfoDb * 1.40.1 2024-05-24 [1] Bioconductor 3.19 (R 4.4.0)
GenomicRanges * 1.56.2 2024-10-09 [1] Bioconductor 3.19 (R 4.4.1)
ggplot2 * 3.5.1 2024-04-23 [1] CRAN (R 4.4.0)
IRanges * 2.38.1 2024-07-03 [1] Bioconductor 3.19 (R 4.4.1)
kableExtra * 1.4.0 2024-01-24 [1] CRAN (R 4.4.0)
limma * 3.60.6 2024-10-02 [1] Bioconductor 3.19 (R 4.4.1)
lubridate * 1.9.4 2024-12-08 [1] CRAN (R 4.4.2)
MatrixGenerics * 1.16.0 2024-04-30 [1] Bioconductor 3.19 (R 4.4.0)
matrixStats * 1.5.0 2025-01-07 [1] CRAN (R 4.4.2)
openxlsx * 4.2.8 2025-01-25 [1] CRAN (R 4.4.2)
purrr * 1.0.2 2023-08-10 [1] CRAN (R 4.4.0)
readr * 2.1.5 2024-01-10 [1] CRAN (R 4.4.0)
S4Vectors * 0.42.1 2024-07-03 [1] Bioconductor 3.19 (R 4.4.1)
SingleCellExperiment * 1.26.0 2024-04-30 [1] Bioconductor 3.19 (R 4.4.2)
SpatialExperiment * 1.14.0 2024-05-01 [1] Bioconductor 3.19 (R 4.4.2)
stringr * 1.5.1 2023-11-14 [1] CRAN (R 4.4.0)
SummarizedExperiment * 1.34.0 2024-05-01 [1] Bioconductor 3.19 (R 4.4.0)
tibble * 3.2.1 2023-03-20 [1] CRAN (R 4.4.0)
tidyr * 1.3.1 2024-01-24 [1] CRAN (R 4.4.0)
tidyverse * 2.0.0 2023-02-22 [1] CRAN (R 4.4.0)
TreeSummarizedExperiment * 2.12.0 2024-04-30 [1] Bioconductor 3.19 (R 4.4.2)
XVector * 0.44.0 2024-04-30 [1] Bioconductor 3.19 (R 4.4.0)
[1] /home/orue/R/x86_64-pc-linux-gnu-library/4.4
[2] /usr/local/lib/R/site-library
[3] /usr/lib/R/site-library
[4] /usr/lib/R/library
──────────────────────────────────────────────────────────────────────────────A work by Migale Bioinformatics Facility
Université Paris-Saclay, INRAE, MaIAGE, 78350, Jouy-en-Josas, France
Université Paris-Saclay, INRAE, BioinfOmics, MIGALE bioinformatics facility, 78350, Jouy-en-Josas, France

---
params:
my.interest: "BU"
title: "RESTORBIOME2: Differential analysis"
subtitle: "Experiment with `r params$my.interest`"
author:
- name: Olivier Rué
orcid: 0000-0003-1689-0557
email: olivier.rue@inrae.fr
affiliations:
- name: Migale bioinformatics facility
adress: Domaine de Vilvert
city: Jouy-en-Josas
state: France
- name: Christelle Hennequet-Antier
orcid: 0000-0001-5836-2803
email: christelle.hennequet-antier@inrae.fr
affiliations:
- name: Migale bioinformatics facility
adress: Domaine de Vilvert
city: Jouy-en-Josas
state: France
date: "2023-05-02"
date-modified: today
#bibliography: ../../../resources/biblio.bib # don't change
#csl: ../../../resources/biomed-central.csl # don't change
# # Do not modify this section without taking precautions
# # Don't remove the commented line, it is usefull for building the site with this new project!
license: "This document will not be accessible without prior agreement of the partners"
format:
html:
embed-resources: false
toc: true
toc-location: right
page-layout: article
code-overflow: wrap
code-fold: true
code-tools: true
code-summary: "Show the code"
---
```{r}
#| label = "setup",
#| include = FALSE
knitr::opts_chunk$set(echo =TRUE,
cache = FALSE,
message = FALSE,
warning = FALSE,
fig.height = 3.5,
fig.width = 10.5)
```
```{r}
#| label = "setuplib"
#Use packages and functions
library(tidyverse)
library(edgeR)
library(ggplot2)
library(kableExtra)
library(ExperimentSubset)
library(colorspace)
library(openxlsx) # manipulation excel files
#sessionInfo()
```
# Load data
We used datasets created at the exploration and normalisation step from `r params$my.interest` experiment. Genes with too low expression were filtered. A normalization factor is calculated to take into account the different sizes of the sequencing banks (i.e. the total read count) and the distribution of reads per sample on sequencing run, as discussed [@Dillies2012-ln]. Read counts were normalized using trimmed mean of M-values (TMM) method [@Robinson2010] from **edgeR** {{< iconify mdi tools >}} R package.
```{r}
#| label = "se",
#| echo = FALSE,
#| results = FALSE
# load a SummarizedExperiment object
# cat("Experiment ", params$my.interest)
se_ofinterest <- paste("../output/Restorbiome_se_", params$my.interest, ".rds", sep="")
se <- readRDS(se_ofinterest)
print(se)
#samples metadata
colData(se)
#add colours
```
```{r}
#| label = "dge"
# load a dge object
dge_ofinterest <- paste("../output/Restorbiome_dge_", params$my.interest, ".rds", sep="")
dge <- readRDS(dge_ofinterest)
# print(dge)
head(dge$counts, n=3)
```
Above an extract from the expression matrix which contains expression on `r nrow(dge$counts)` expressed genes and `r ncol(dge$counts)` samples from `r params$my.interest` experiment.
# Differential analysis
The aim is to identify the genes differentially expressed between treatment and Glucose.
The differential analysis is based on a Generalized Linear Model (GLM) to take into account the distribution of count-type data (non-normality of the data) and over-dispersion. The regression coefficients of the factors are estimated by a log-linear model.
Using **edgeR** {{< iconify mdi tools >}} R package, the GLM model [@McCarthy2012-ij] was fitted with one factor of two levels:
* control condition: Glucose (G)
* treatment: Raffinose (R)
```{r}
#| label = "Design"
dge$samples$group <- as.factor(dge$samples$group)
dge$samples$group <- relevel(dge$samples$group, ref = paste(params$my.interest, "G", sep="_"))
# levels(dge$samples$group)
#matrix design: one factor group
design <- model.matrix(~ 0 + group, data = dge$samples)
colnames(design) <- gsub(paste0("group", params$my.interest, "_"), "", colnames(design))
# design
```
```{r}
#| label = "Contrasts"
#--- make all contrasts of interest
# treatment versus control condition
RvsG <- makeContrasts(R-G, levels=colnames(design))
my.contrasts <- list(RvsG = RvsG)
names(my.contrasts) <- paste(params$my.interest, names(my.contrasts), sep="_")
```
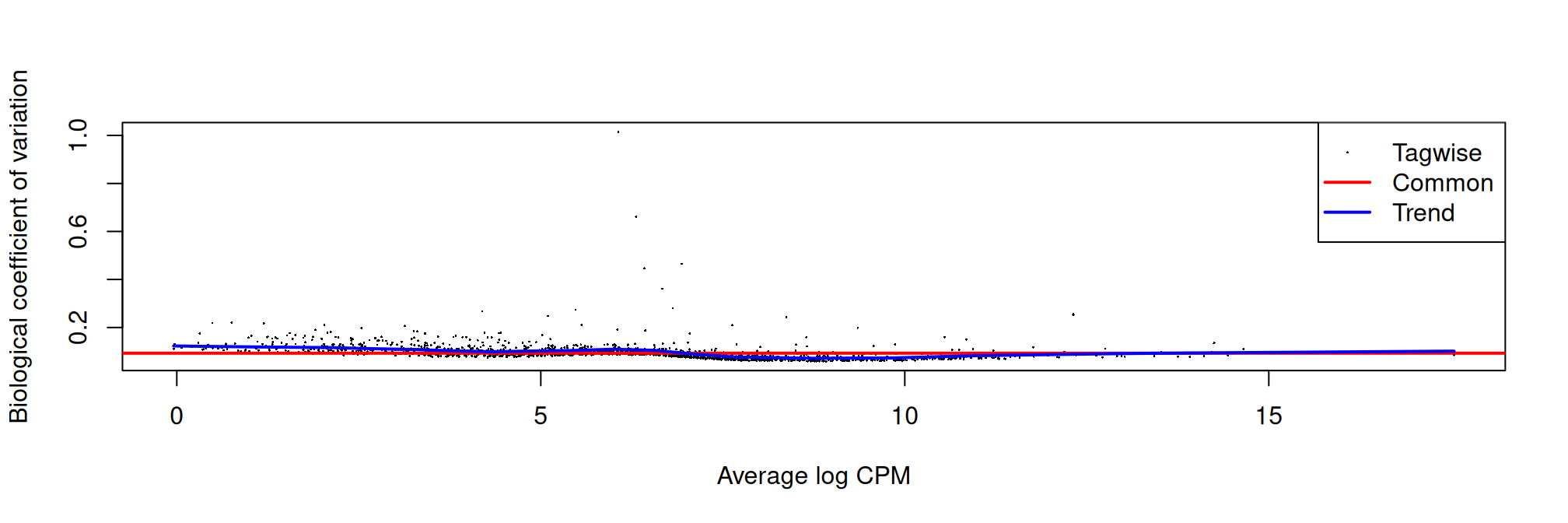
We set robust=TRUE when estimating the dispersion. This has no effect on the downstream analysis,
but is nevertheless very useful for identifying outliers from the trend of the mean dispersion of the negative binomial distribution.
```{r}
#| label = "Dispersion"
# robust at gene level
# with specific design
dge<-estimateDisp(dge, design = design, robust=TRUE)
#summary(dge$tagwise.dispersion)
plotBCV(dge)
```
We used GLM model [@McCarthy2012-ij] to perform likelihood ratio tests for each contrast defined as treatment versus Glucose control treatment. List of differentially expressed genes were obtained after controlling the false discovery rate with a Benjamini-Hochberg correction [@Benjamini1995] at 0.05 threshold.
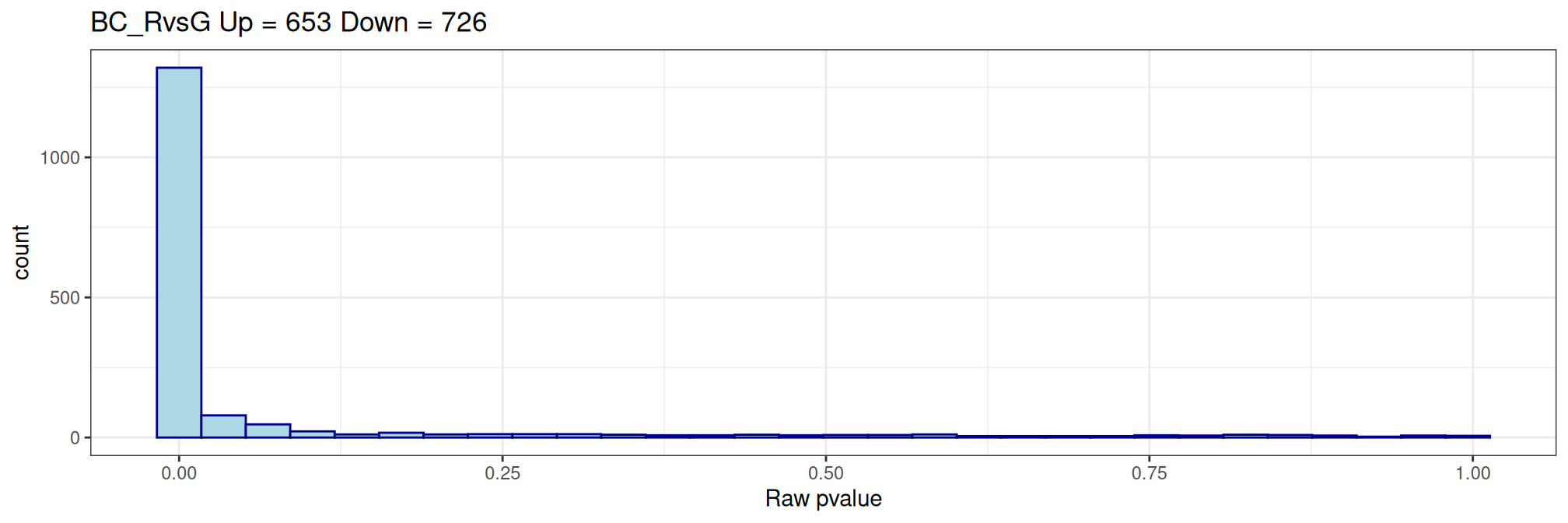
Histogram of raw p-values and mean-difference plot (also called MA-plot) are useful graphs to validate the model.
```{r}
#| label = "Diff"
#only the trended dispersion is used under the QL
method <- "LRT"
#fit <- glmFit(dge, dge$design)
# with specific design
fit <- glmFit(dge, design = design)
#head(fit$coefficients)
#head(fit$dispersion)
res<-list()
nDiffTotal <- NULL
#fit model for each contrast
for (ctr in 1:length(my.contrasts)){
qlf<- glmLRT(fit, contrast = my.contrasts[[ctr]])
topTags(qlf)
# Results, Nb of DE
#cat("contrast= ",names(my.contrasts)[ctr],"\n")
#print(summary(decideTestsDGE(qlf)))
nDiffTotal <- cbind(nDiffTotal, summary(decideTestsDGE(qlf)))
# results of tests
res[[ctr]]<-topTags(qlf,n=nrow(dge$counts),adjust.method="BH",sort.by="none")$table
names(res)[ctr]<-names(my.contrasts)[ctr]
# histogram of raw pvalues
my.title <- paste(names(my.contrasts)[ctr], "Up =", nDiffTotal[,ctr]["Up"], "Down =", nDiffTotal[,ctr]["Down"])
p <- ggplot(data = qlf$table, aes(x = PValue))
p <- p + geom_histogram(color="darkblue", fill="lightblue")
p <- p + theme_bw() + xlab("Raw pvalue")
p <- p + ggtitle(my.title)
print(p)
# MA plot
plotMD(qlf, main = my.title)
}
# results combined in a list object
# initialisation
#--- summary number of DE by contrast
nDiffTotal <- as.data.frame(nDiffTotal)
colnames(nDiffTotal) <- names(my.contrasts)
cat("\nNumber of features down/up and total:\n")
nDiffTotal %>% kbl() %>% kable_styling() %>% scroll_box(width = "100%", height = "200px")
# write.table(nDiffTotal,
# file = paste("../output/2_Differential_Analysis",params$my.interest,".resume.xls", sep=""),
# sep = "\t", row.names = TRUE,
# dec = ".", quote = FALSE)
```
```{r}
#| label = "Save",
#| echo = FALSE
my.exportResults.edgeR <- function (out.edgeR, group, counts, annot, Idannot, alpha = 0.05, export = TRUE) {
dge <- out.edgeR$dge
res <- out.edgeR$results
tmm <- dge$samples$norm.factors
N <- colSums(dge$counts)
f <- tmm * N/mean(tmm * N)
normCounts <- round(scale(dge$counts, center = FALSE, scale = f))
base <- data.frame(Id = rownames(counts), counts)
names(base) <- c("Id", colnames(counts))
norm.bm <- data.frame(Id = rownames(normCounts), normCounts)
names(norm.bm) <- c("Id", paste0("norm.", colnames(normCounts)))
norm.bm$baseMean <- round(apply(scale(dge$counts, center = FALSE,
scale = f), 1, mean), 2)
for (cond in levels(group)) {
norm.bm[, cond] <- round(apply(as.data.frame(normCounts[,
group == cond]), 1, mean), 0)
}
base <- merge(base, norm.bm, by = "Id", all = TRUE, sort=F)
complete <- list()
for (name in names(res)) {
complete.name <- base
res.name <- data.frame(Id = rownames(res[[name]]), FC = round(2^(res[[name]][,
"logFC"]), 3), log2FoldChange = round(res[[name]][,
"logFC"], 3), pvalue = res[[name]][, "PValue"], padj = res[[name]][,
ifelse("FDR" %in% names(res[[name]]), "FDR", "FWER")])
complete.name <- merge(complete.name, res.name, by = "Id",
all = TRUE)
dge.add <- data.frame(Id = rownames(dge$counts), tagwise.dispersion = round(dge$tagwise.dispersion,
4), trended.dispersion = round(dge$trended.dispersion,
4))
complete.name <- merge(complete.name, dge.add, by = "Id",
all = TRUE)
complete.name <- merge(annot, complete.name, by.y = "Id", by.x = Idannot, all = TRUE, sort=F)
complete[[name]] <- complete.name
if (export) {
up.name <- complete.name[which(complete.name$padj <=
alpha & complete.name$log2FoldChange >= 0), ]
up.name <- up.name[order(up.name$padj), ]
down.name <- complete.name[which(complete.name$padj <=
alpha & complete.name$log2FoldChange <= 0), ]
down.name <- down.name[order(down.name$padj), ]
# using openxlsx package to export results
excelsheets <- list(complete = complete.name,
up = up.name,
down = down.name)
write.xlsx(excelsheets, file = paste0("../html/",
name, ".xlsx"))
}
}
return(complete)
}
# exporting results of the differential analysis
out.edgeR<-list(dge = dge, results = res)
complete <- my.exportResults.edgeR(out.edgeR, counts = dge$counts, group = dge$samples$group, annot = dge$genes, Idannot = "Gene_ID", alpha=0.05)
```
```{r}
#| label = "saveResults_DE",
#| echo = TRUE
# List of DE result from each contrast as a dataframe with informations like logFC, PValue, FDR...
saveRDS(res, file=paste("../output/Restorbiome_DE_", params$my.interest,".rds",sep=""))
```
# Reproducibility token
```{r}
#| label = "sessionInfo"
sessioninfo::session_info(pkgs = "attached")
```
# References